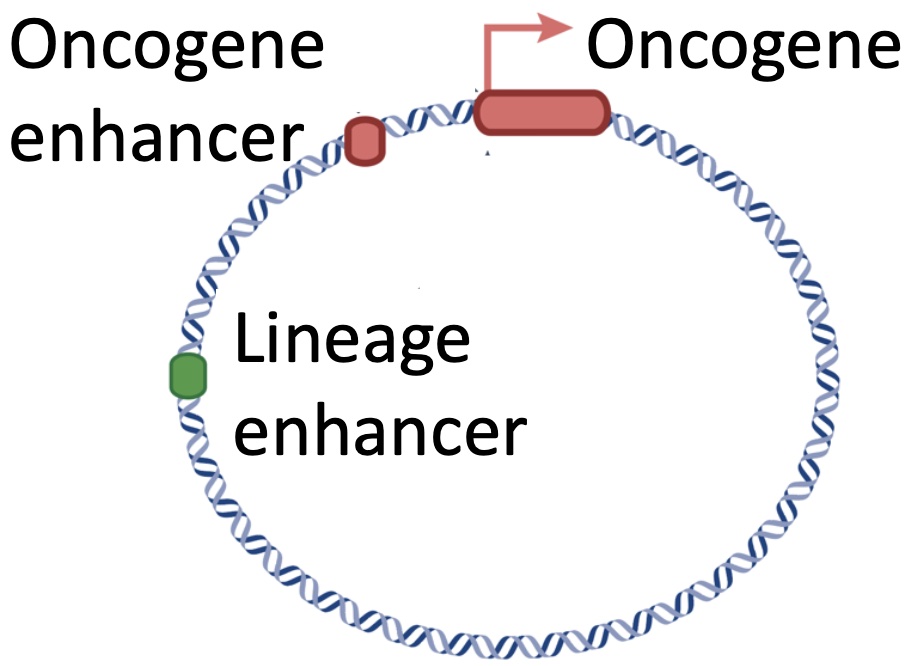
Most cancer genomes experience DNA amplifications as the most common gain-of-function alterations leading to overexpression of oncogenes.
Why does this work? How does the transcriptional regulation of the oncogenes get hijacked on these amplifications? Do they exploit tissue-specific enhancers and transcription factors for oncogene expression and cancer growth?
We combine whole genome sequencing of hundreds of breast and lung cancers including long-read and epigenetic analyses with validation of our findings using CRISPR-based genome engineering to answer these questions.
Understanding which regulatory factors are essential for cancer growth could lead to novel tissue-specific gene regulation targeting therapies.
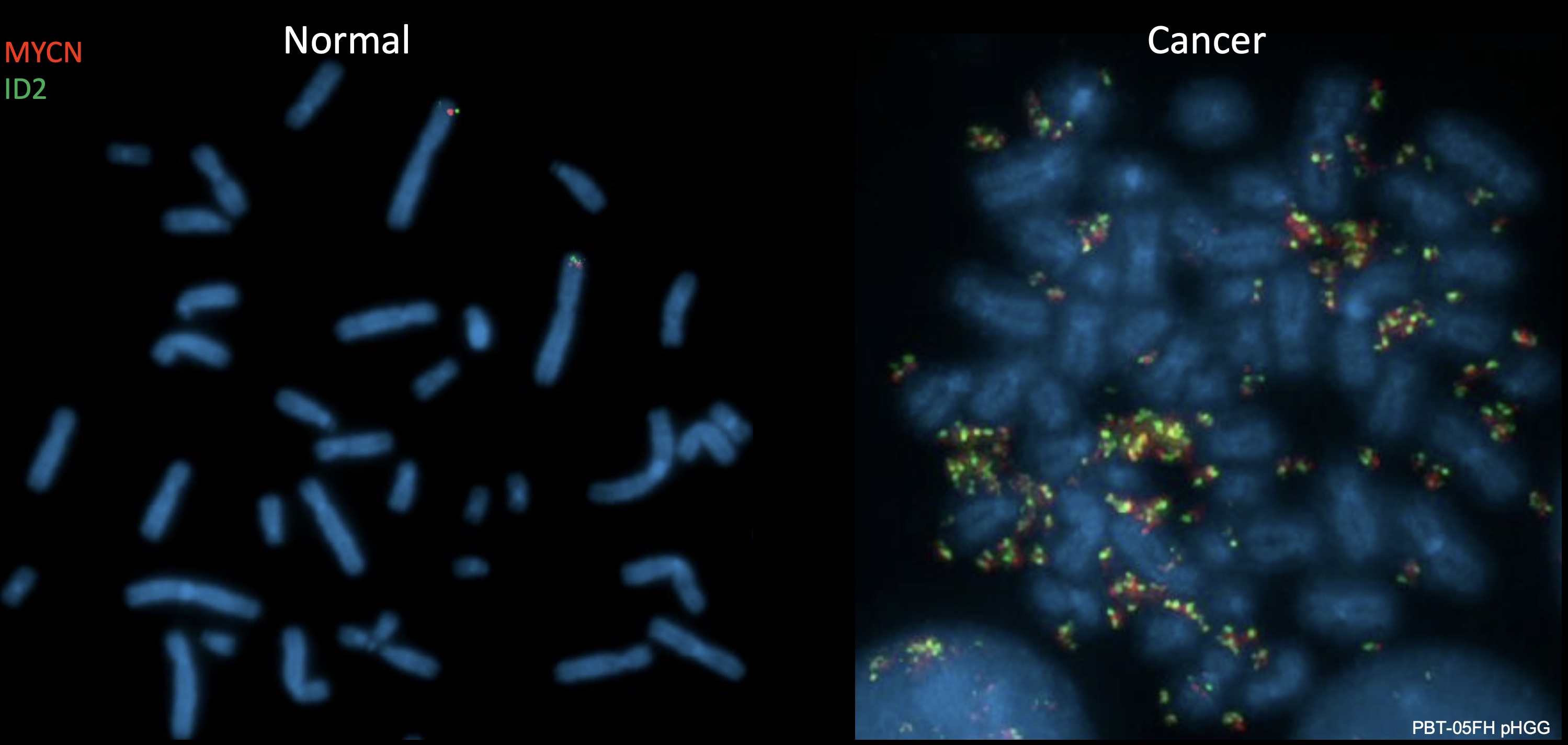
Metaphase FISH image of cells with normal karyotype (left) and brain cancer cells with a high-level amplicon of MYCN and ID2 on extrachromosomal DNA (right) – a phenomenon only observed in malignant cells. (image from Dubois F et al. Nature Cancer. 2022.)
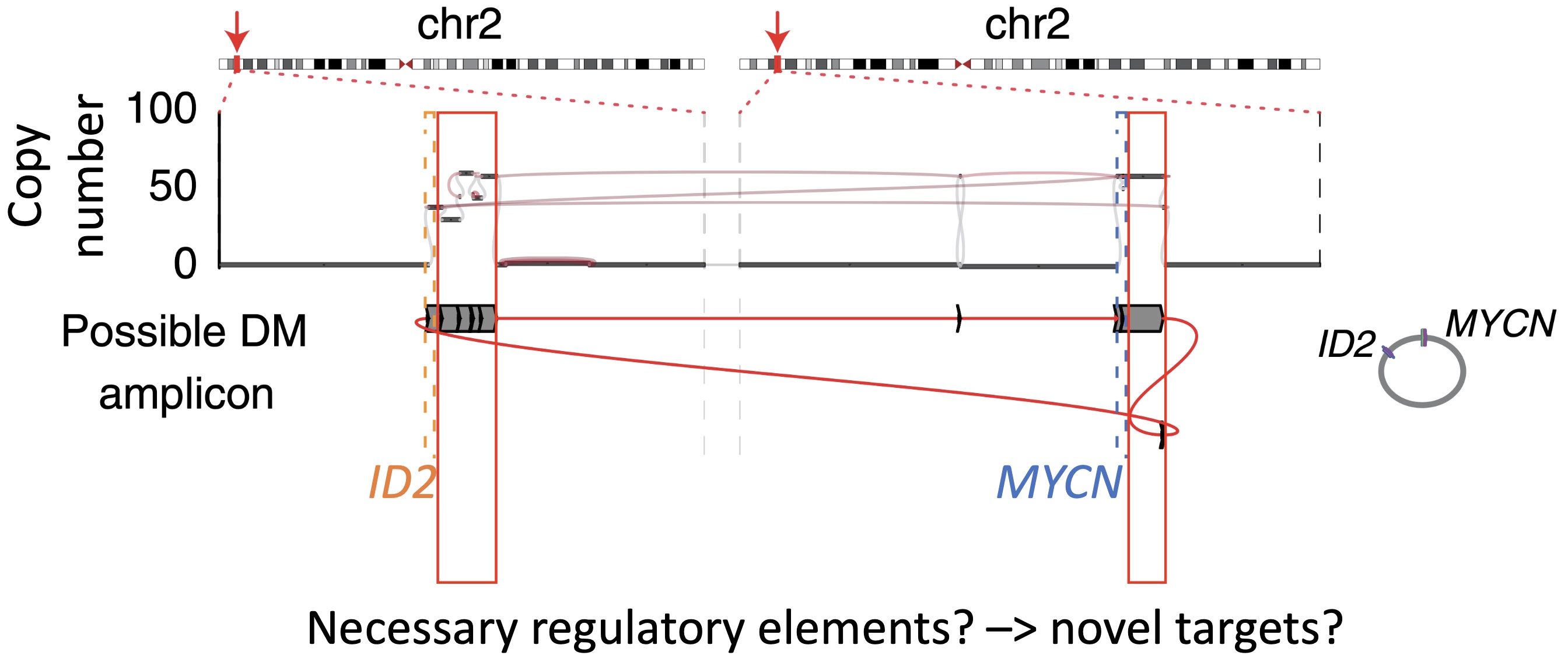
The coding sequences of the oncogenes are only a small fraction of the amplified DNA. Why are the cancer cells keeping 50+ copies of the rest? Are there essential regulatory elements in these co-amplified regions?
Reconstruction of an MYCN-ID2 extrachromosomal DNA (ecDNA) amplicon. The plot shows two loci on chromosome 2 with the positions of the coding sequences of the MYCN and ID2 genes highlighted in blue and orange respectively. The larger red boxes show co-amplified regions that could contain essential regulatory elements. (from Dubois F et al. Nature Cancer. 2022.)
To answer these questions, we will analyze several hundred breast and lung cancer tumor samples by whole genome sequencing (WGS), including with long-read technology.
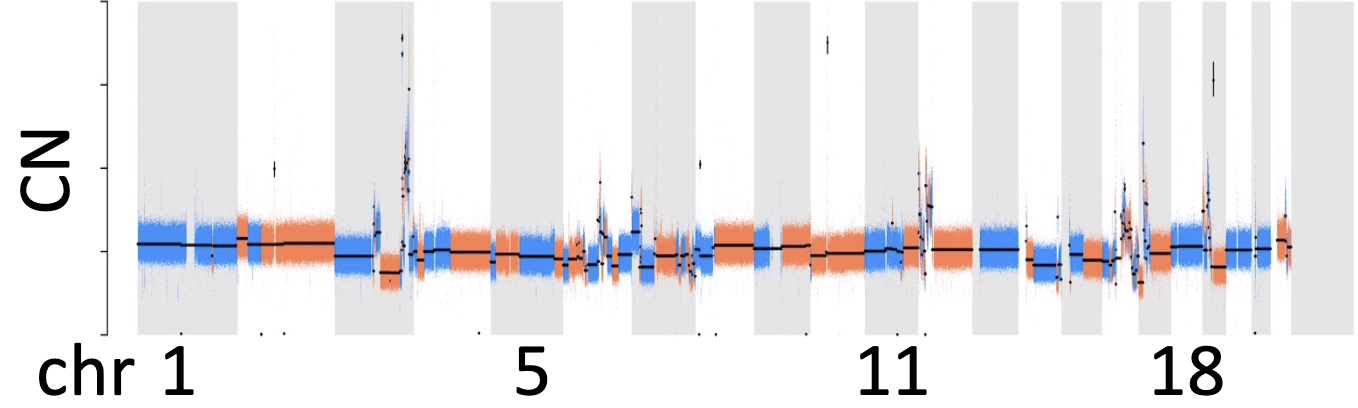
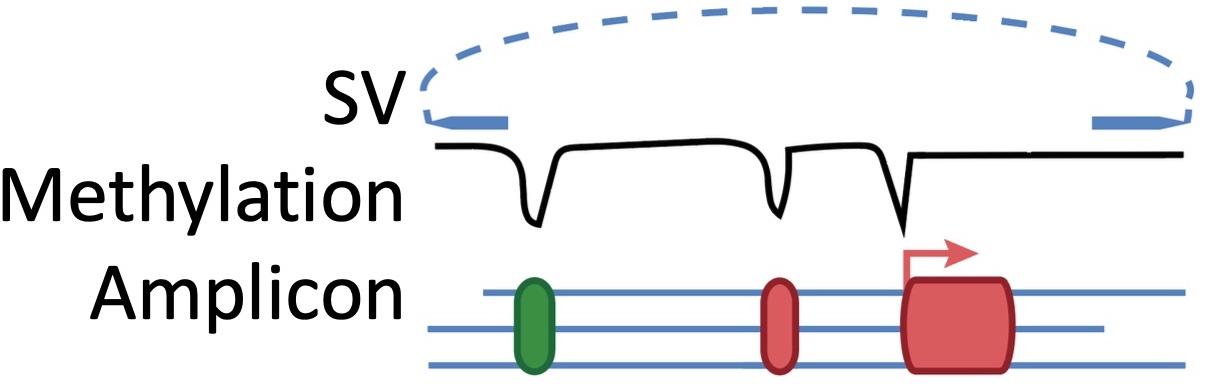
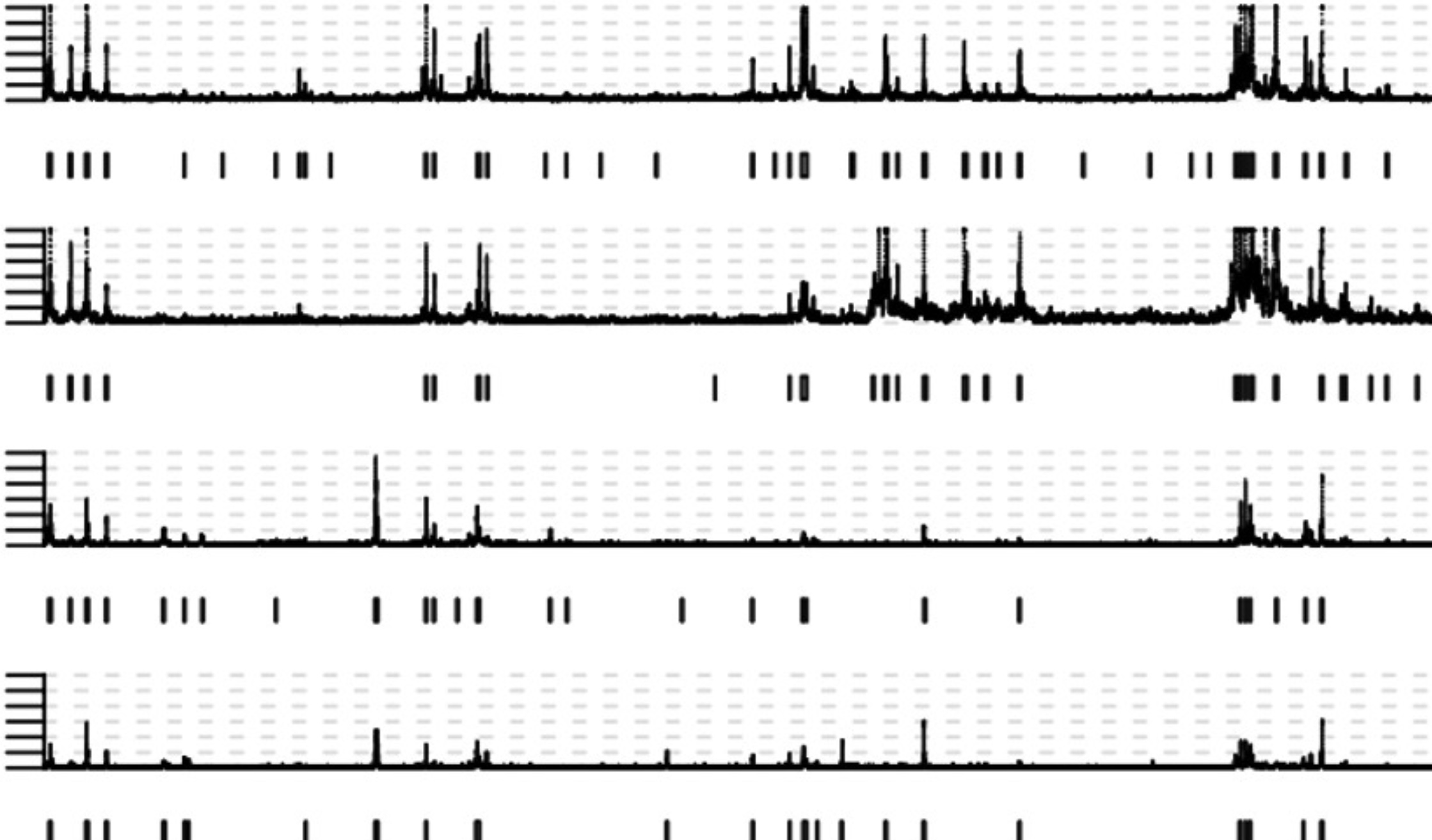
(Left) Copy number profile resulting from WGS. (Center) Long-read technology will allow us to detect SVs in the cancer genome (top track) and methylation (middle track) from which we can infer the positions of enhancers and transcription start sites (bottom track). Combing all this data, we can reconstruct the topology of the amplicon (Right)
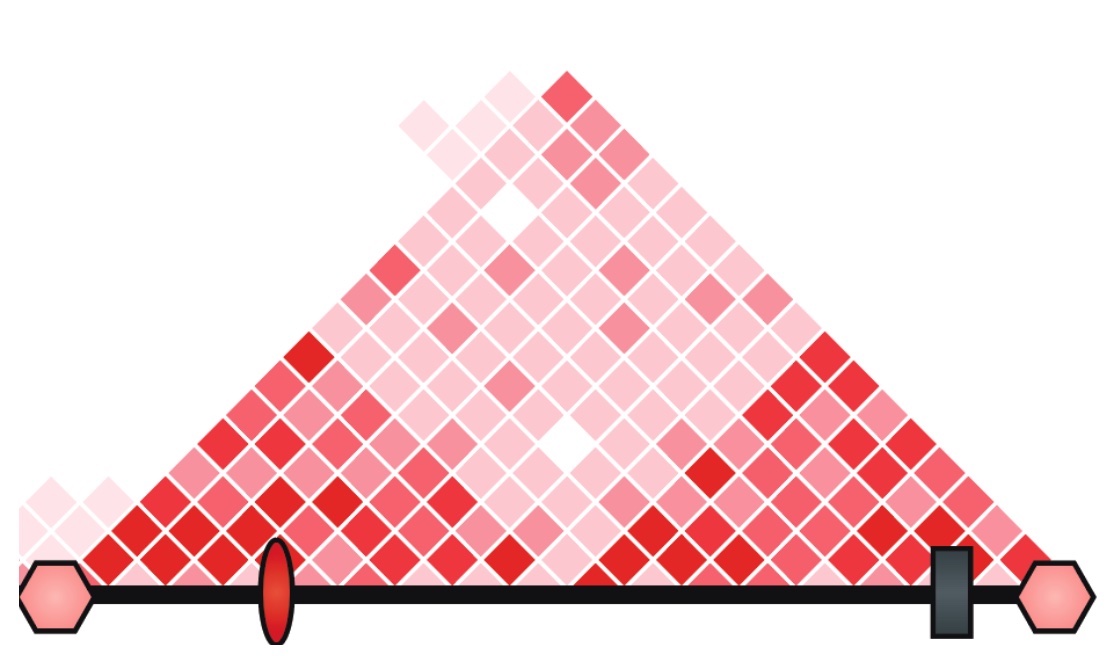
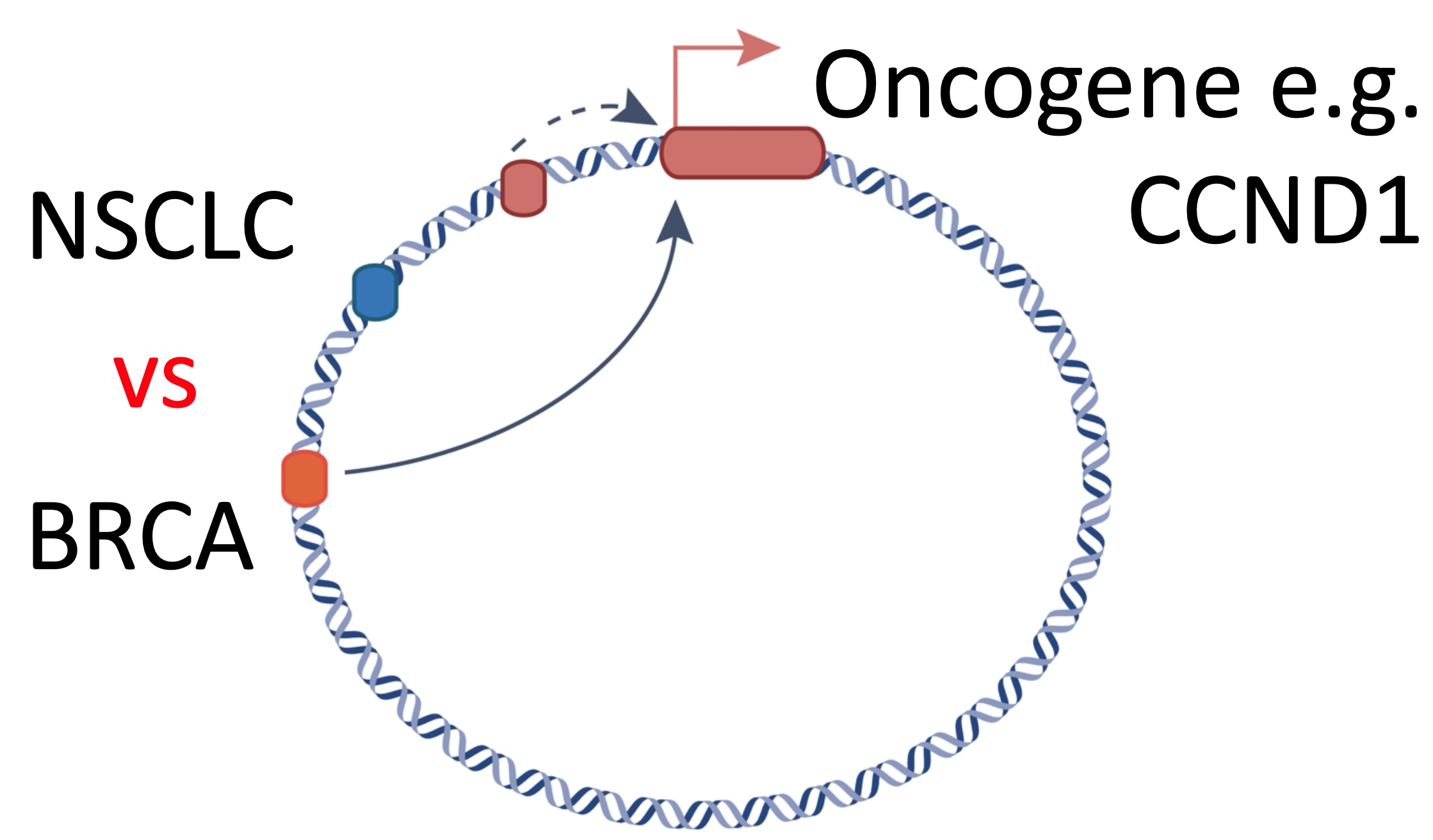
Using differences in the epigenetic profiles (left) between tumors with high (top 2 tracks) and low (bottom 2 tracks) expression of the oncogene and HiC to measure 3D genome interactions (center), we will infer which enhancers on the amplicon activate oncogene transcription. By analyzing amplicons of the same oncogene in both non-small cell lung cancer (NSCLC) and breast cancer (BRCA), we can test if there is tissue type specific enhancer usage (right).
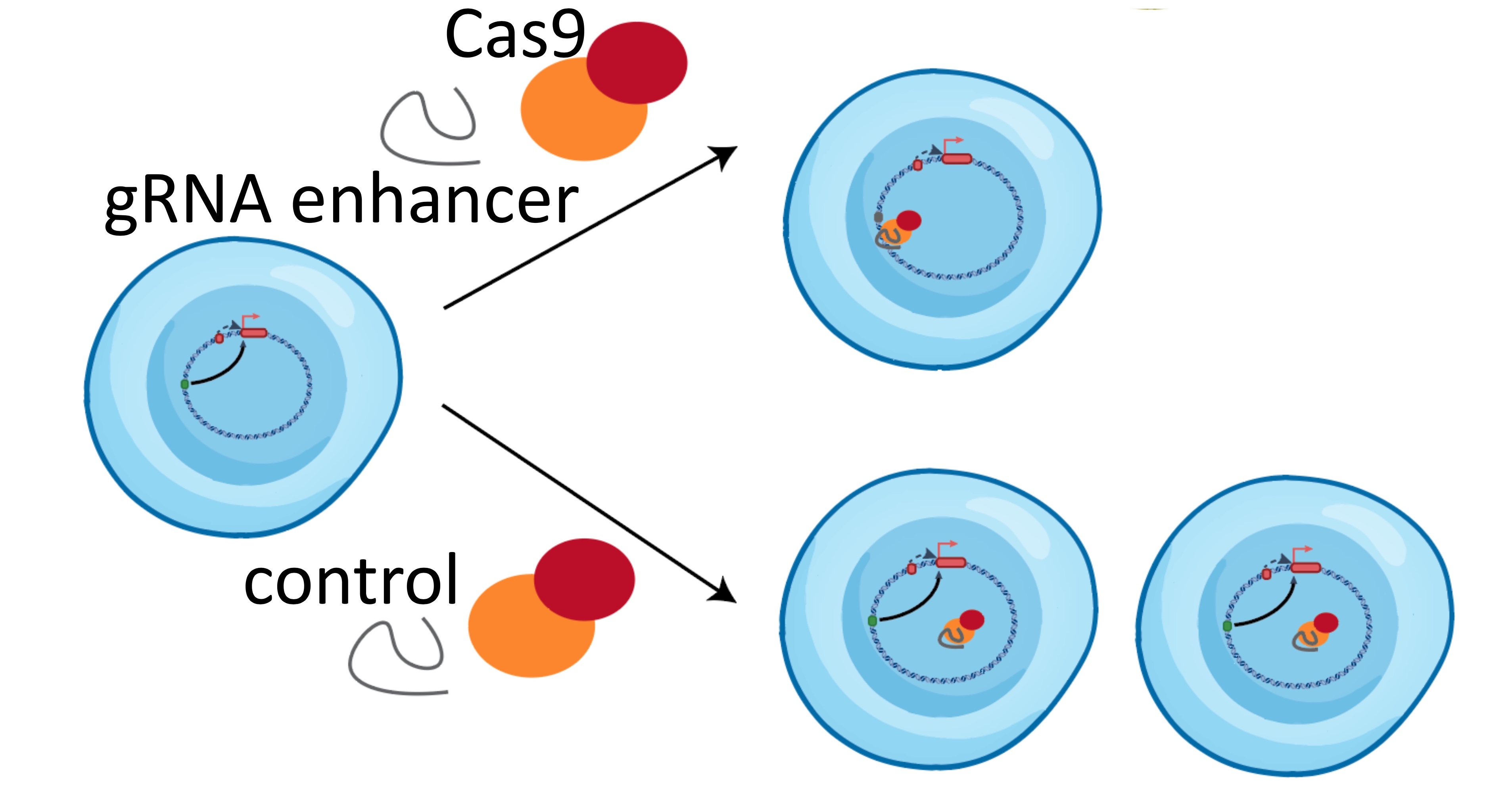
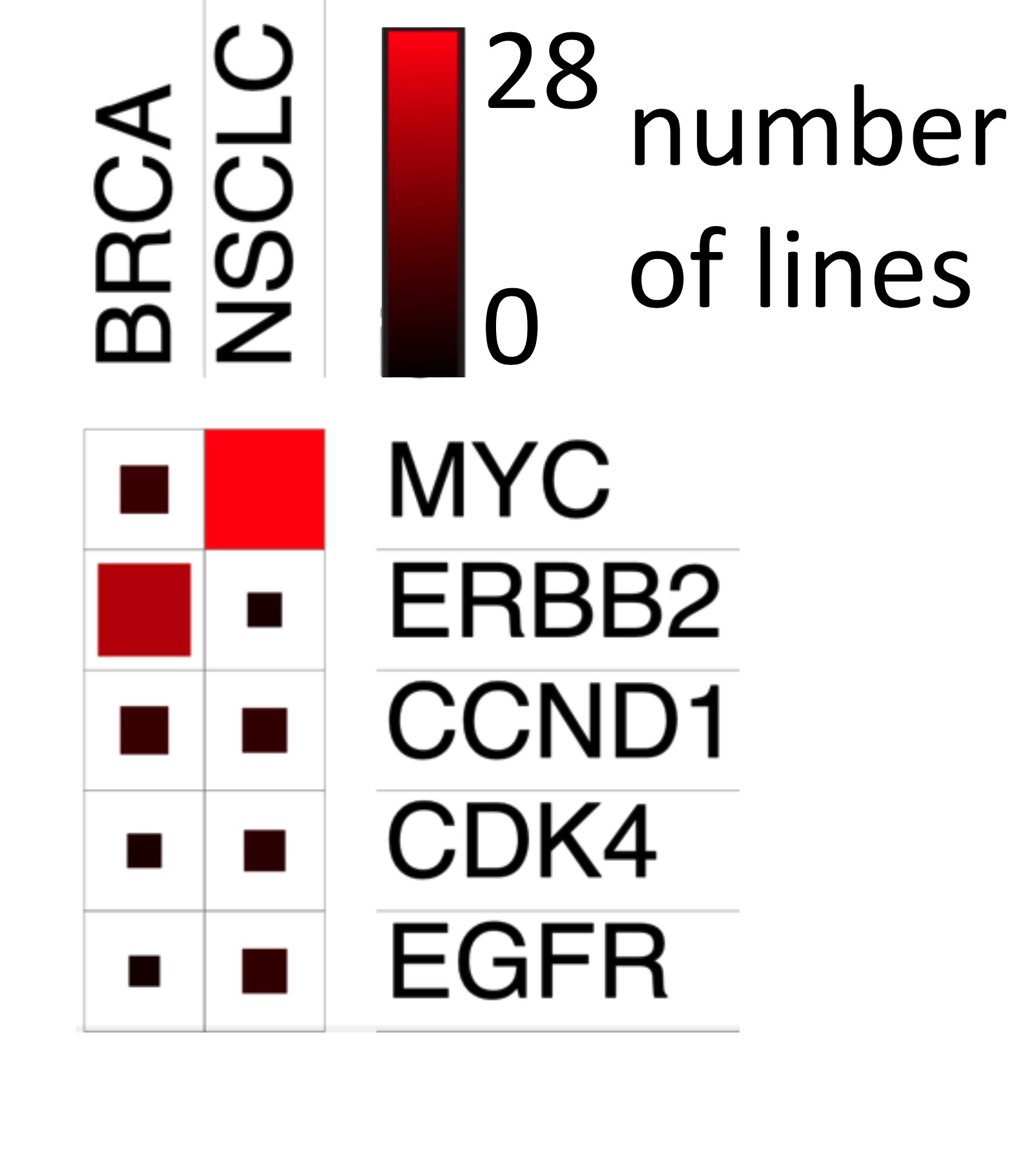
We will validate which regulatory elements are essential for oncogene expression and cancer cell growth using CRISPR-based genome engineering (left). We have already identified dozens of suitable cell lines of both cancer types that show amplifications of the same oncogenes to test our hypotheses (right).
We are part of a multi-institutional network of collaborators combining efforts to understand cancer genomes.
Our collaborators at Charité and other leading cancer centers.